It is a bit of time I was thinking that this post may be of
help to people intersted in genealogical network reconstruction under the
median joining algorihtm. www.Fluxus-engineering.com
provides, free of charge, the phylogenetic Network software in its as of today version
4.611. Network makes use of
the median joining algorithm to generate genealogical trees from DNA,
aminoacid, linguistic and other data but can also provide age estimates for the
ancestral nodes in the tree topology. I have used several times this software
in the past and I was impresed for its versatility, the graphics (compared to
TCS or Splitstree)
and its friendly interface. However, contrary to TCS and Splitstree, the Network team suggests
the use of “recommended” add-ons such as DNA Alignment, Network Publisher and pibase for importing
data without errors, create publication quality graphics and generating network
input files (.rdf) from next generation sequencing files respectively. Despite Network is freely distributed, the add-ons are
not.
In this post I suggest a quick and free mode to create nexus input files for Network starting from DNA or amino acid sequences aligned with… your preferred algorithm. I found this path to be always successful and literally based on few clicks of your mouse but I am sure there are other personalized ways to do it. Example alignment files are from Poore and Andreakis 2012.
1)
Align your sequences and create a fasta file; pay attention
to the taxon names: the first 6 characters need to be different
2)
Import
fasta file or paste sequences into this great format conversion online tool: the format converter
v2.0.5
3)
Convert
file into “nexus sequential” format
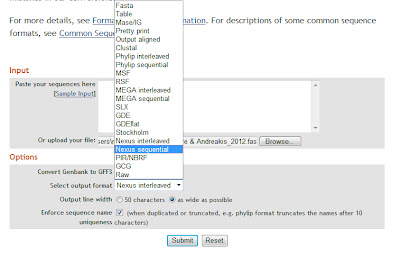
5) Open the nexus
file in WordPad and edit the nexus lines as follows:
From:

To:

Semplified results:



Thank you! Your how-to helped saved my evening and finally generated a network. David, CH
ReplyDeleteNo worries david - happy I have been of help:)
DeleteThank you for posting this! It was very helpful. -Verena
ReplyDeleteMy pleasure Verena!
ReplyDeleteI have been trying to format my data correctly to input to network and have been having trouble the past few days. This finally helped me clear up my issues. Thank you so much for this! I'm sure others who are struggling setting up data for Network without paying a dime out of their pocket will benefit from this. -Nadia
ReplyDeleteThanks for your nice words and your comment Nadia - I wouldn't mind to spend my $$$ on a fully functional software and then benefit from some of its freely available tools; but not the other way around. Very bad strategy.
DeleteI tried following your suggestion exactly and tried to input my file, but it keeps giving me error messages. Very frustrating! They have made it very tricky. This software is a trap.
DeleteNope my friend, it is really a piece of cake. I am happy to try for you if you send my your input file.
Deletebest
Niko
thanks alot Nikos! It is very helpful
ReplyDeleteThis great site has been very helpful but when I was still having problems Nikos kindly had a look at my sequence file. He explained that I had a return between the sequence name and the sequence where there should be a space. Thanks so much for your help!
ReplyDeleteThanks Nikos!! This is really helpful.I was fascinated to see this tips as I was searching for a way to tackle network. but a problem arose.After importing edited nexus sequential file to network, it says " sequence 1 doesn't have the specified no. of nucleotides.". What is your advice over this? Thank you
ReplyDelete-Pamoda
Hi Pamoda and many thanks for your email.
DeleteWhat is likely happening in this case is the following:
You have initially imported your sequences in Bioedit to create your alignment but you haven't exported your fasta file as alignment; you have simply saved it as a fasta file.
In the latter case, your sequences, although aligned manually, are of different length because of missing data in their end. If the file is only saved as fasta and not exported as alignment, Bioedit will not match the differences of the end missing data in length by adding the final gaps. The results is that once you have created your nexus file via the path I suggest, some sequences will be shorter that the length of the alignment and therefore they will not have the specific number of nucleotides reported in the nexus block.
Quick solution: If you do not want to repeat the full path, simply open your nex file in notepad and you will likely (in the end of your alignment) find that some of your sequences are shorter than others usually by one or few gaps. You will see that the gasp are missing. Just add the gaps manually in notepad, save and your input file is ready for use in network.
Please let me know if this worked for you and feel free to send me over your fasta file to check it out.
best
Nikos
Hi Nikos,
ReplyDeleteYa, it worked. I implemented your quick solution and I got it. So the next time, I should give the aligned file to format converter.? Is that what you meant? However , I started making a fresh input file likewise and once I download the nexus sequential formated file from format converter, it automatically saves with.txt extention though I make it .nexus and it is not opened in network, sorry to trouble you oe after another.
Thank you
-Pamoda
Hi Nikos, I hope you are still checking this blog. I've selected the Median-joining method and followed your steps, but when I try to load my file, I get the error message "This appears to be a multistate rdf file. The reduced median network can only accept binary rdf files. Please apply the median--joining network method instead, or recode your file as binary rdf file'. Do you have any suggestions?
ReplyDeleteHi my friend, I assume, once you have followed my suggestions for file preparation you have tried to load you file as a nexus file.
DeletePlease let me know otherwise please send me over your file in fasta format and I will be happy to give it a go
best
N
Yes, I followed your suggestions and I am still having problems. Do you have an e-mail to which I can send you my files? I have what should be a fairly simple analysis, but I can't seem to make the file load correctly (I've also tried converting it using DnaSP, and I've tried using popART to draw my network instead). Something must be wrong with my file(s) but I don't know what
Deletemy email
Deleten.andreakis@aims.gov.au
Thanks! (I've sent you my data.)
DeleteHi Nikos, I just wanted to thank you again for your help. You are a life-saver!
Deletemany thanks for your message my friend, I am so glad I have been of help!
Deletei ahve followed the same alignment in bioedit but net work still give an error I've selected the Median-joining method and followed your steps, but when I try to load my file, I get the error message "This appears to be a multistate rdf file. The reduced median network can only accept binary rdf files. Please apply the median--joining network method instead, or recode your file as binary rdf file'. Do you have any suggestions?
ReplyDeleteI have the same problem :( and have no idea what to do...
DeleteHi my friend.
DeleteIt is really very easy. First save your data in Bioedit as a fasta ALIGNMENT (saving as an alignment is very important) and then follow the instructions.
Otherwise as with Pamoda, open in wordpad and see if there are missing gaps in the end of your data and fill them.
In any case if you still have problems please send me over your data and I will be happy to check them out
Best
N
Dear Nikos,
ReplyDeleteI repaired fasta from Mega and followed your instructions to get input to Network. But I have a message: " sequence 1 doesn't have the specified no. of nucleotides." I tried to resolve this problem with suggestions you gave Pamoda but it didn't :( and the input seems to be ok. I use Network 4.6. Any idea? HEEEELP
Beata
Hi my friend.
ReplyDeleteIt is really very easy. First save your data in Bioedit as a fasta ALIGNMENT (saving as an alignment is very important) and then follow the instructions.
Otherwise as with Pamoda, open in wordpad and see if there are missing gaps in the end of your data and fill them.
In any case if you still have problems please send me over your data and I will be happy to check them out
Best
N
Dear Nikos,
ReplyDeleteThanks a lot for your willing to help. I drew finally my network :) The problem was with my fasta file (prepaired in MEGA). The easiest way was to use Bioedit and all palyed. You were right: "saving as an alignment is very important" :) well... in fact its really easy. Thanks Nikos !!!
Beata
Dear Nikos,
ReplyDeleteThanks a lot for your willing to help. I drew finally my network :) The problem was with my fasta file (prepaired in MEGA). The easiest way was to use Bioedit and all palyed. You were right: "saving as an alignment is very important" :) well... in fact its really easy. Thanks Nikos !!!
Beata
Dear Nikos,
ReplyDeleteThank you so much for this tutorial. However, I am having a problem. I started out using MacVector to align my sequences and then saved it in FASTA format and followed the instructions. However, I am not getting an error that says "The maximum number of characters (10000) is attained" and the program freezes. Please let me know if you have a solution.
-Meg
Hi Nikos,
ReplyDeleteThank you for the wonderful tutorial. However, I am having an issue. I aligned my sequences using MacVector, saved them as a FASTA alignment, and then followed your instructions. But now I am getting an error which states "The maximum number of characters (10000) has been attained." and then the program promptly freezes and the error window will not disappear. Please let me know if you have a solution!
-Meg
Hey Meg, thanks for your kind message. This process works with any alignment algorithm I have used so far. At this stage your error message can originate from any small typo in the file. Open your converted nexus file and check it out. Otherwise send it over to me and I can quickly check it for you.,
Deletebest
N
I just sent you the file in an email.
DeleteDear Nikos,
Deleteyour suggestions in this blog were very helpfull in importing data into network.I have problem interpreting my network.please help me in this regard
hi bro, can you explain me how draw reduced median network.
ReplyDeleteIt helps me a lot, thank you very much!!!
ReplyDeletedear Nikos
ReplyDeletei did as you said but network doesnt import nexus file. how do i import that
Dear Nikos,
ReplyDeleteAre you aware of any free/shareware software that woud digest Network (tree/network) output format and being able to translate it to PDF or any other useful vector format for free? Paying 145$ just to get one PDF out of Network Publishes is just a bit too much for a cup of average coffee. Thanks a lot, Tine
I want through your comment too fast mate, the answer is yes:
DeleteopART 1.7 (http://popart.otago.ac.nz/downloads.shtml). Extraordinary user friendly with great graphic ability and the capacity to geotag or colour code your data on a build-in map. Apologies, N
cheers
N
Hi TiNe,
ReplyDeletePle4ase send over a link of your software and I will be happy to try it out.
FYI, I have recently came across popART 1.7 (http://popart.otago.ac.nz/downloads.shtml). Extraordinary user friendly with great graphic ability and the capacity to geotag or colour code your data on a build-in map
cheers
N
Hi Nikos,
ReplyDeleteI have been getting the same error message that some of the other commenters have: "This appears to be a multistate rdf file. The reduced median network can only accept binary rdf files. Please apply the median--joining network method instead, or recode your file as binary rdf file"
I have followed your instructions and tried fixing it myself with no luck. Is there any chance I could send you my files to have a look at?
Kind regards,
Tahlia
Hey Tahlia, please feel free to do so and I will try to be of help.
Deletecheers
n
Hi Nikos
ReplyDeleteThanks a lot for this great help. BUt I would like to ask, what to do in case my first 6 characters are not different?
Gaelle
Hi Gaelle,
DeleteI usually ad a number followed by taxon name like this
1_taxonname
2_taxonname
3_taxonname
etc.
You can quickly edit these lines in EXCEL.
Cheers
Niko
Hello, Nikos!
DeleteThank you very much for this page.
There was a PDF file on the page, but now only " Error (404) We can't find the page you're looking for. "
And the pictures on the page has a very bad resolution. I can not view which the lines in *nexus file i need to delete to obtain one for Network software.
Could you please send me these pictures in better resolution?
Thank you very much!
Oleg
Hey Oleg,
DeleteI will try to fix it.
Please send me your email and I will forward the files
Cheers
niko